@ Bionanoscience and Biochemistry laboratory, Małopolska Centre of Biotechnology, Jagiellonian University

OVERVIEW
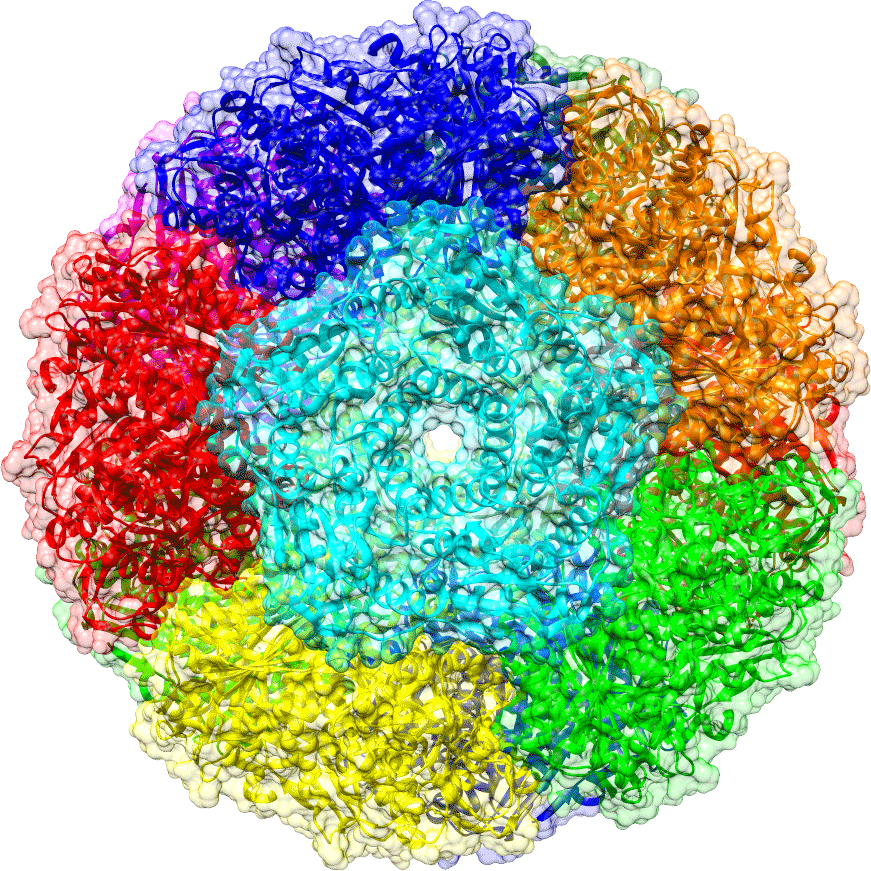
Protein cages are naturally occurring nano- to micro-size hollow particles formed entirely by hierarchical protein self-assembly. This protein shell can provide a distinct environment for guest molecules in biological complexity, of which strategy is widely employed for diverse purposes across different species. Protein cages have also been extensively exploited in the laboratory as ideal platforms for constructing delivery/display vehicles, reaction chambers, and novel nanomaterials. Our research team seeks to uncover the protein cage’s potentials as powerful tools for molecular and synthetic biology. We are preliminarily focusing on a cage-forming lumazine synthase and exploiting and expanding the protein cages through redesign and directed evolution. Namely, current projects are aimed (i) to develop a method for directed evolution of proteins; (ii) to establish a new biotechnology for heterologous protein production; and (iii) to understand the theory underlying the hierarchical assembly for future artificial compartment designs. We anticipate that our protein cage-based biotechnologies will contribute significantly to the development of interdisciplinary research realms ranging from basic biophysics, chemistry to therapeutics.